Methylation analysis of whole genomes and targeted regions
Investigators: Christopher Schröder, Marcel Bargull, Michelle Zechner & Sven Rahmann
Collaborators: Bernhard Horsthemke (Human Genetics); formerly the DEEP consortium
Funding: internal; formerly BMBF (DEEP project)
Software: CAMEL
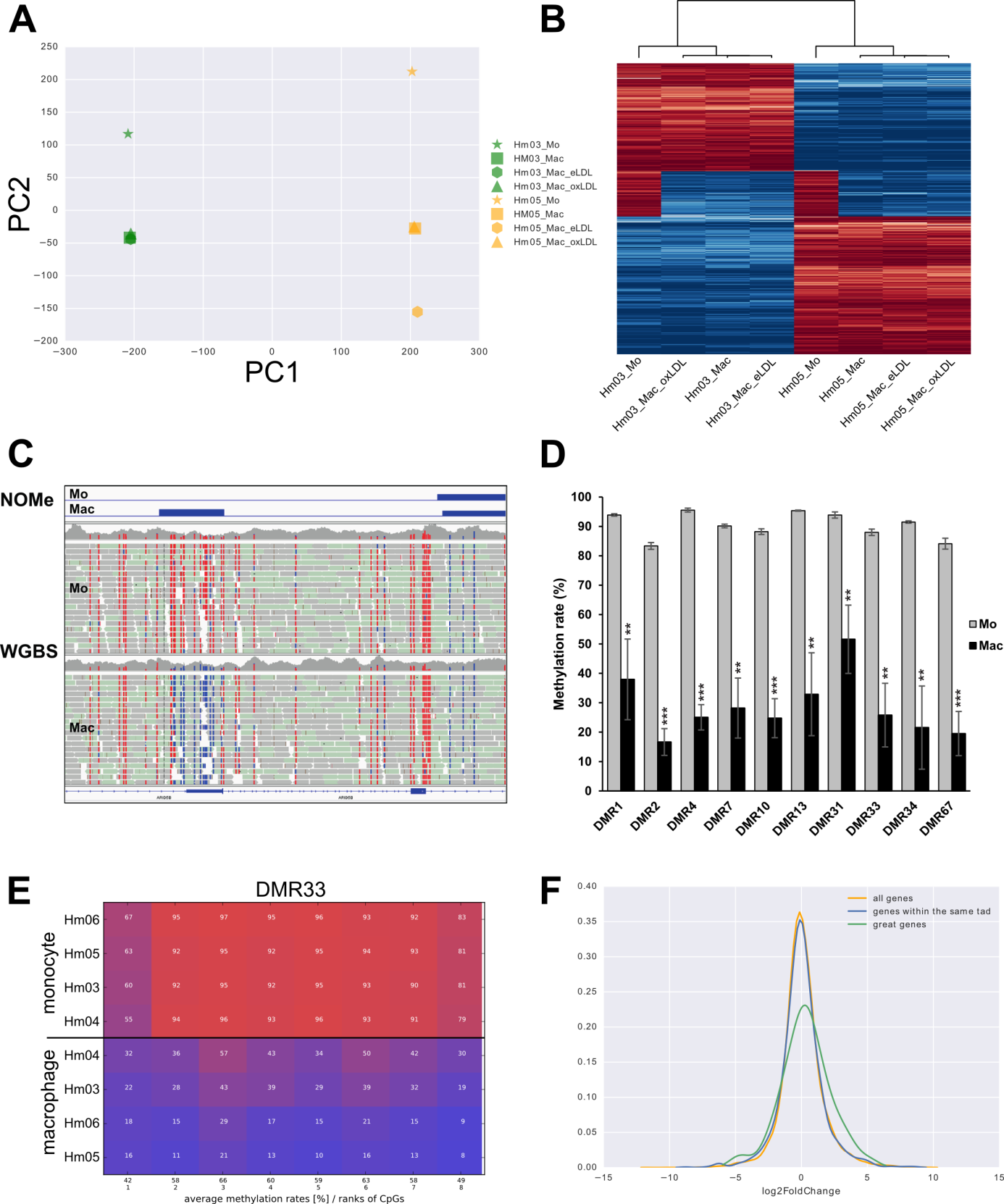
We develop novel principled methods to discover differentially methylated regions (DMRs) in different classes of samples from whole genome bisulfite sequencing data. We also develop analysis and visualization tools for targeted bisulfite sequencing like amplikyzer2.
Christopher Schröder, in cooperation with the Institue of Human Genetics, was part of the “German Epigenome Programme” (DEEP). The project focused on the analysis of cells connected to complex diseases with high socio-economic impact. DEEP contributed epigenome research data to the ‘International Human Epigenome Consortium’ (IHEC) by generating and interpreting up to 80 reference epigenomes of selected human cells and tissues.
Our research focused on the detection of methylation differences from whole genome bisulphite sequencing (WGBS) and “Nucleosome Occupancy and Methylome sequencing” (NOMe-seq). We correlated the differences to other genetic and epigenetic data generated by DEEP e.g. SNPs, long RNA, short RNA, mRNA or ChIP.
In particular, we investigated the epigenetic dynamics of monocyte to macrophage differentiation, where we discovered a phagocytic gene network that is repressed by DNA methylation in monocytes and rapidly de-repressed after the onset of macrophage differentiation.
During the analysis of the differentiation of monocytes and macrophages, we also observed major differences in the methylation between monocytes of different donors. Our current main focus is directed towards these interindividual methylation differences. We found that such differences correlate with a single proximal SNP in more than 1200 samples.
Publications
- S. Wallner, C. Schröder, E. Leitão, T. Berulava, C. Haak et al. Epigenetic dynamics of monocyte-to-macrophage differentiation. Epigenetics & Chromatin 9:33, 2016. doi:10.1186/s13072-016-0079-z.
Algorithmic Bioinformatics, SIC, Saarland University | Privacy notice | Legal notice



